Uploading Your Data
This tutorial teaches you how to format and upload a dataset. With an uploaded dataset, OpenProtein.AI can visualize your data and train models to optimize your designs.
What you need before getting started
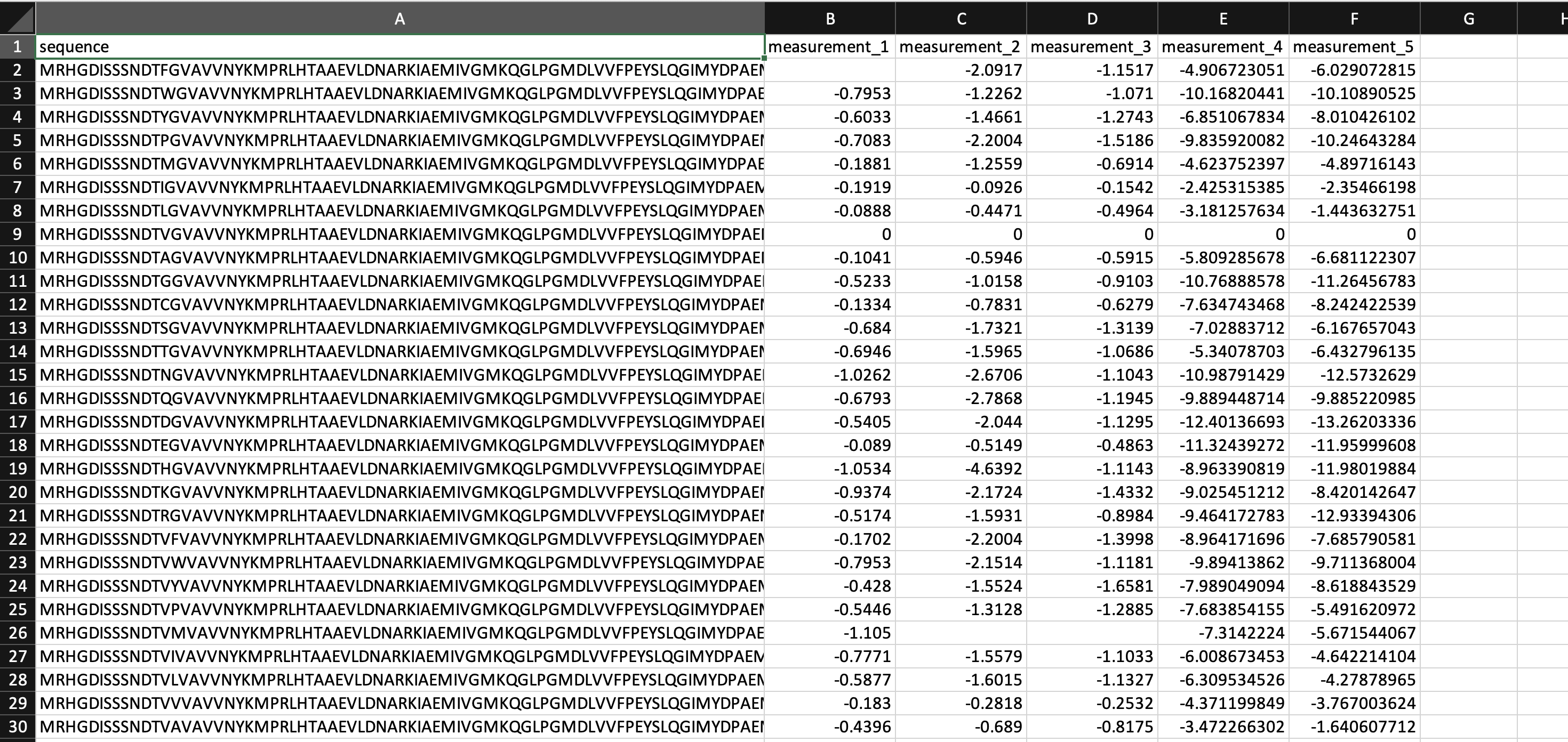
Format your dataset as a CSV table with columns containing: * the full sequence or mutation code of each variant. * measurement values.
Mutation codes denote the specific substitutions that occur at a site. For example, A20T would indicate that alanine at site 20 is substituted with threonine. You can specify multiple mutation codes per sequence by using commas between mutations.
Use a hyphen (-) to indicate each gap in your sequence. Missing measurements are acceptable.
Creating a project and uploading your data
Create a separate project for each parent sequence you use. If you are signing in for the first time, follow the prompt on your screen. You can also select New Project on the main project page.

Start by naming your project and including an optional description.
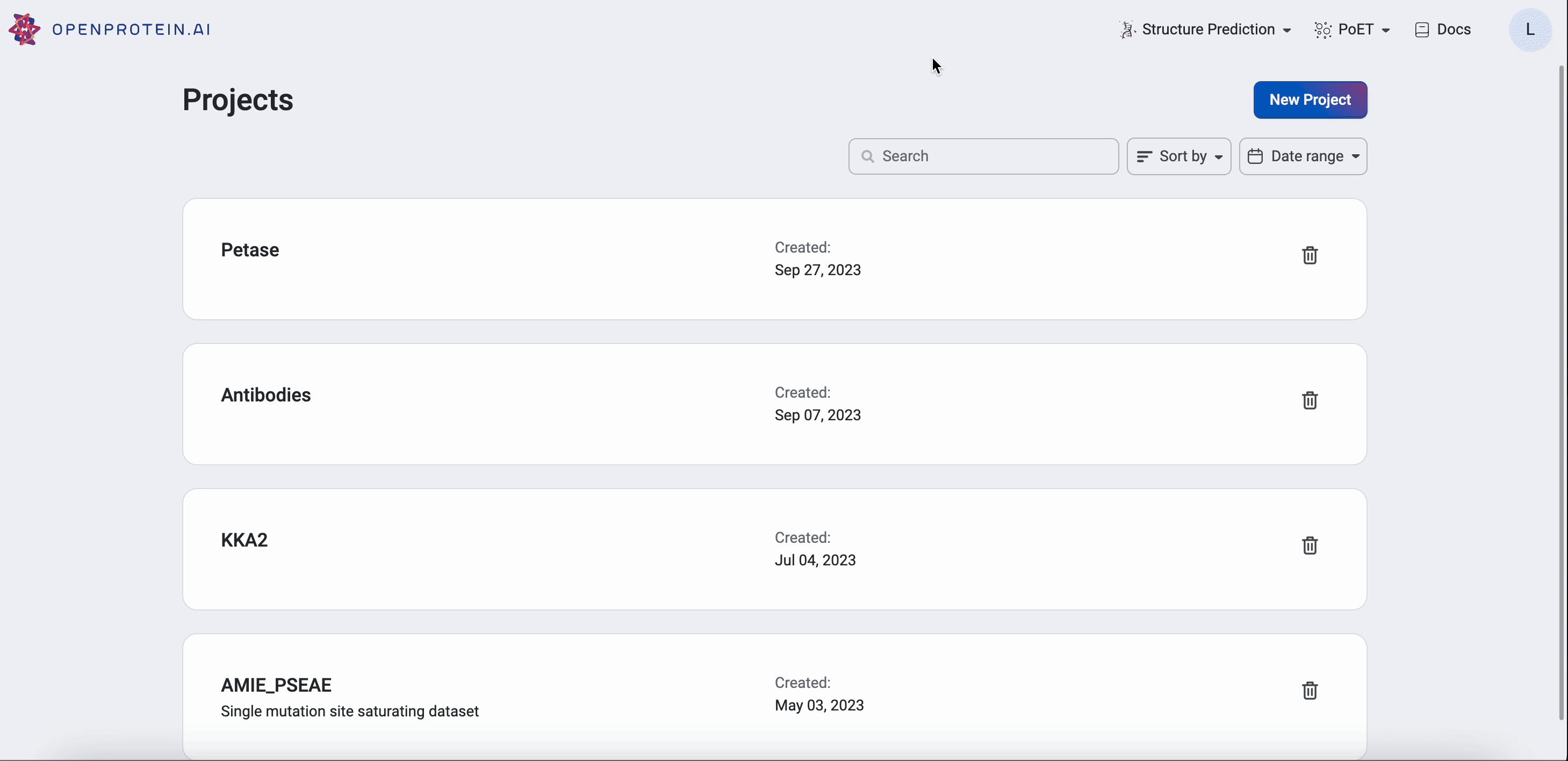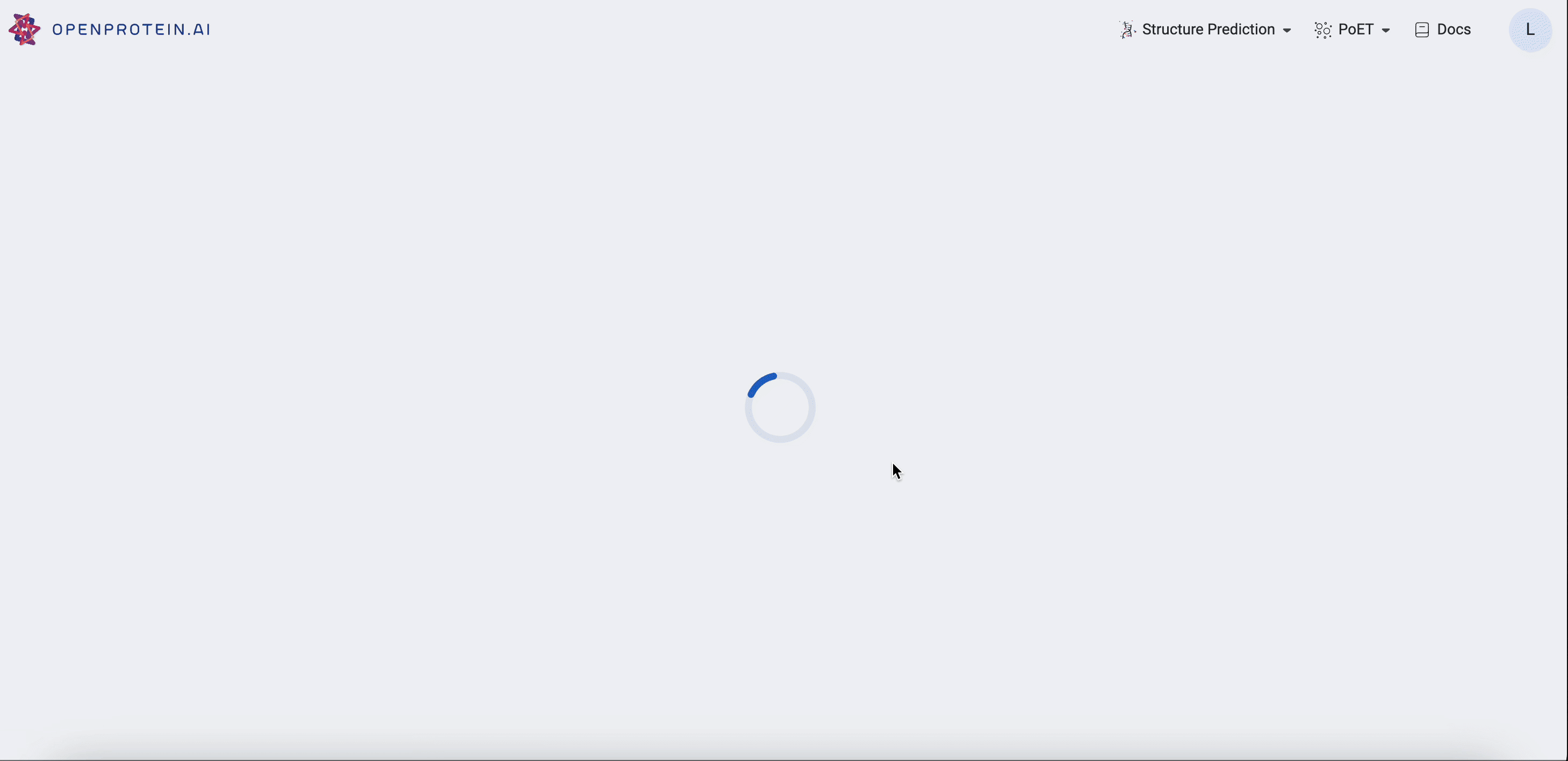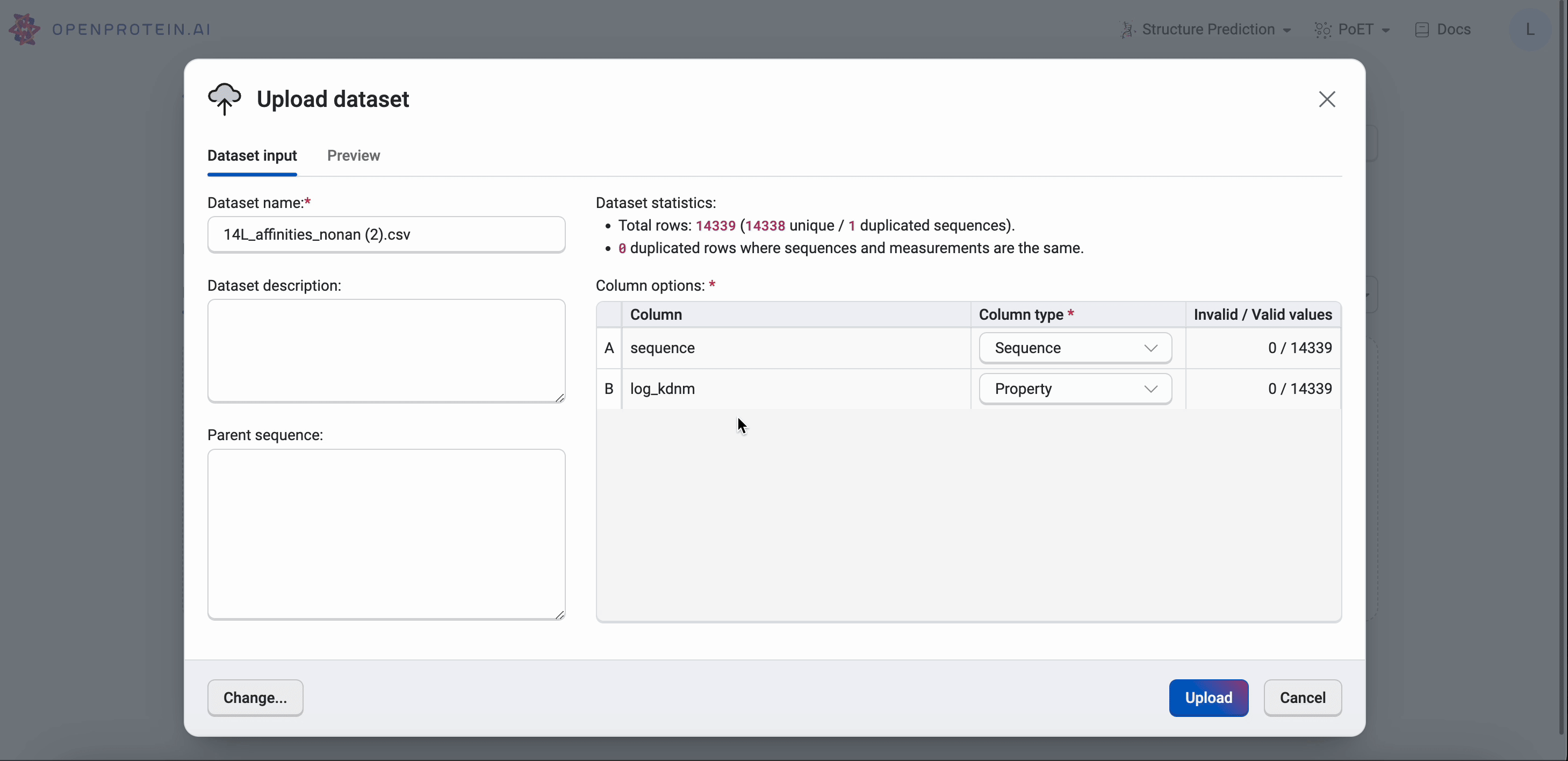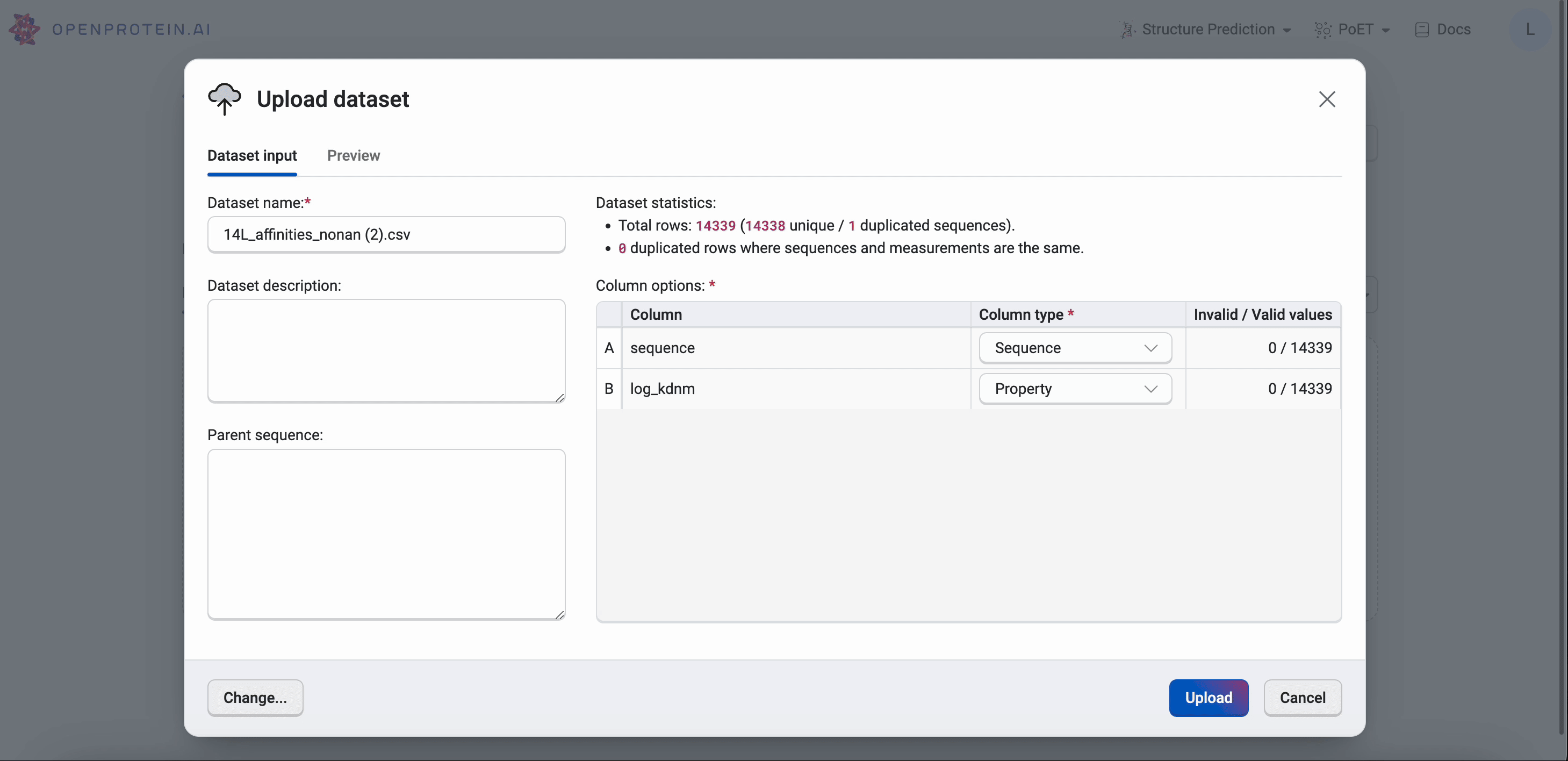
Select Upload dataset and navigate to your CSV file in the file explorer. You can enter a new name for the dataset, or use the default file name. You can also include a description of the dataset, or select a different file using the Change button.
After selecting Open, the application automatically detects your sequence’s columns based on the column name.
If the column types are not automatically filled, you can input them manually using the dropdown menus. The following options are available: * Identifier: user assigned names for each sequence (this is subjective, and dependent on what the user decides to name the sequence). * Property: measurements corresponding to the property that user wish to optimize. 1 property should be 1 column. * Sequence: the sequences of your variants * Mutant: mutation codes. If you use mutation codes, you will be required to input the parent sequence. * Ignored: sequences will not be uploaded.
For datasets containing sequences with mutation codes, enter the wild-type sequence of your protein in the Parent sequence field, then select mutant in the Column type dropdown menu. The platform will process and display the full sequence of each variant specified by your mutation codes.
Select Upload when you are ready to upload your dataset.
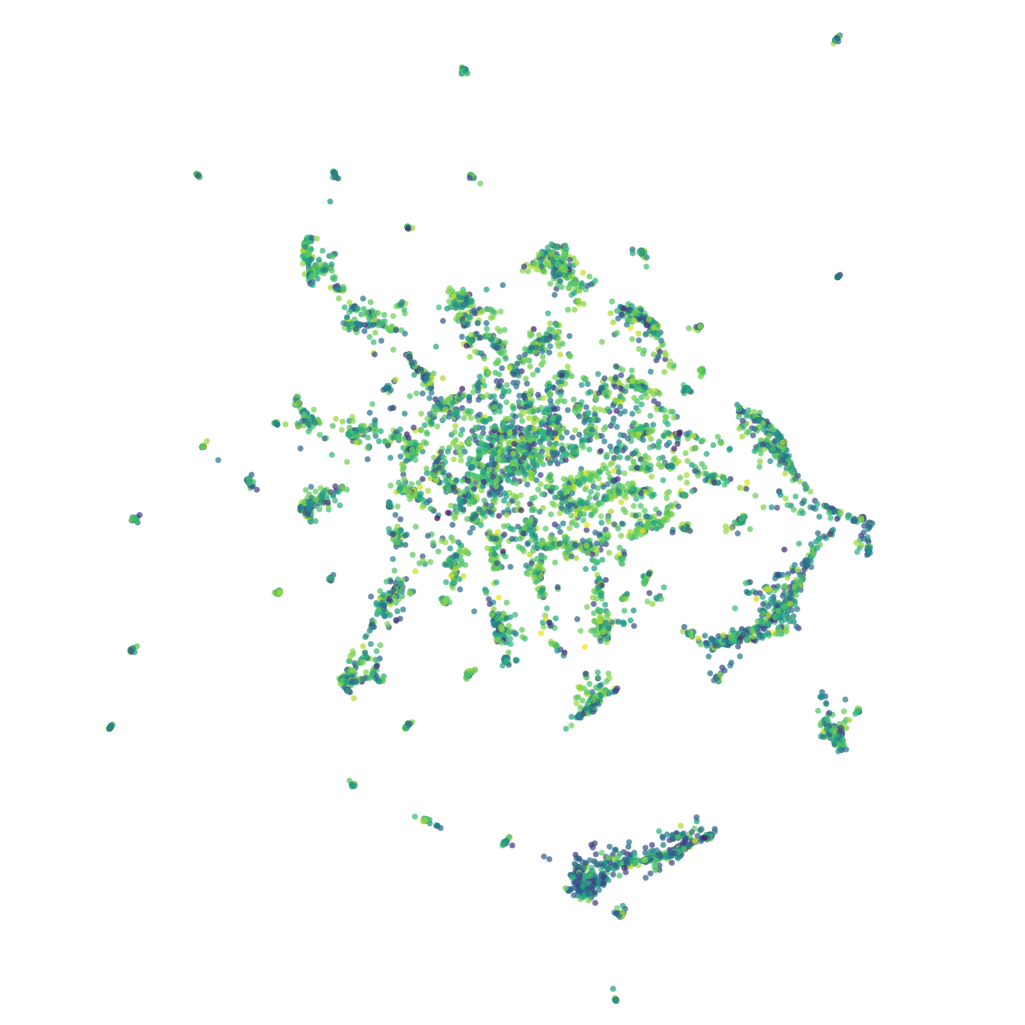
Visualizing your data
Each point in the UMAP visualization represents a single sequence variant.
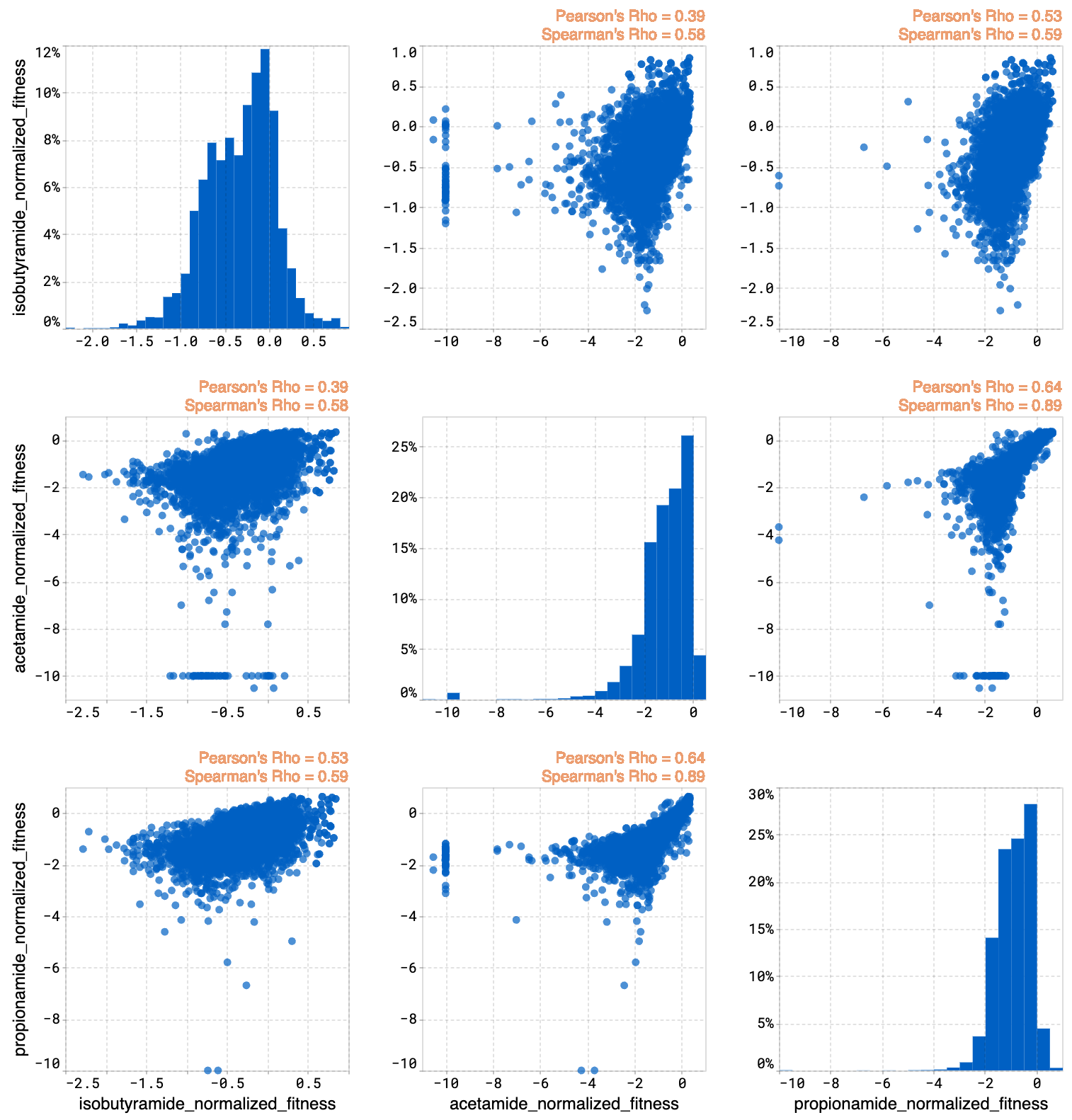
The joint plot displays the pairwise relationships between properties from your dataset. You can visually explore your data by selecting properties and viewing the distribution of individuals of each variable.
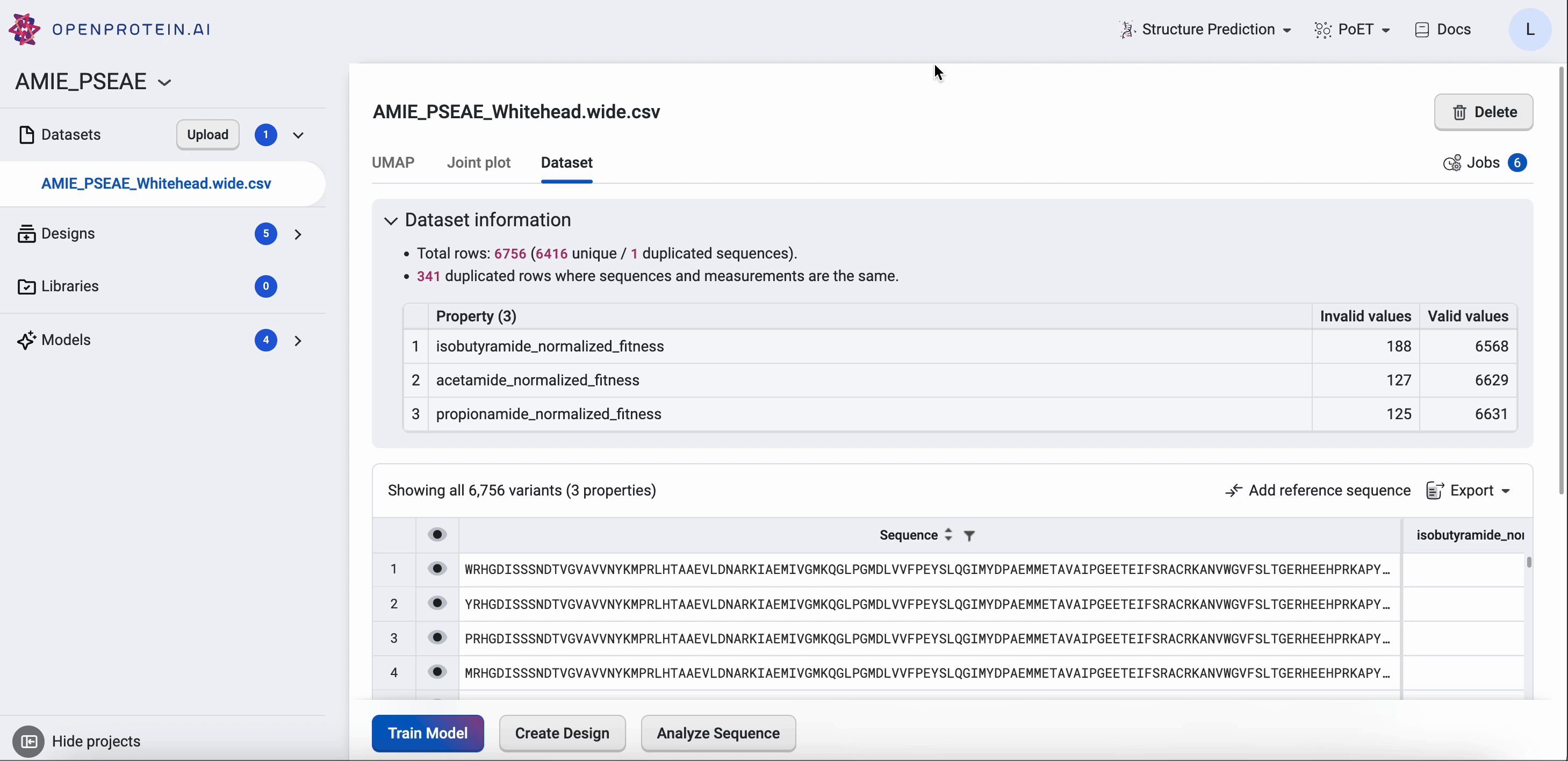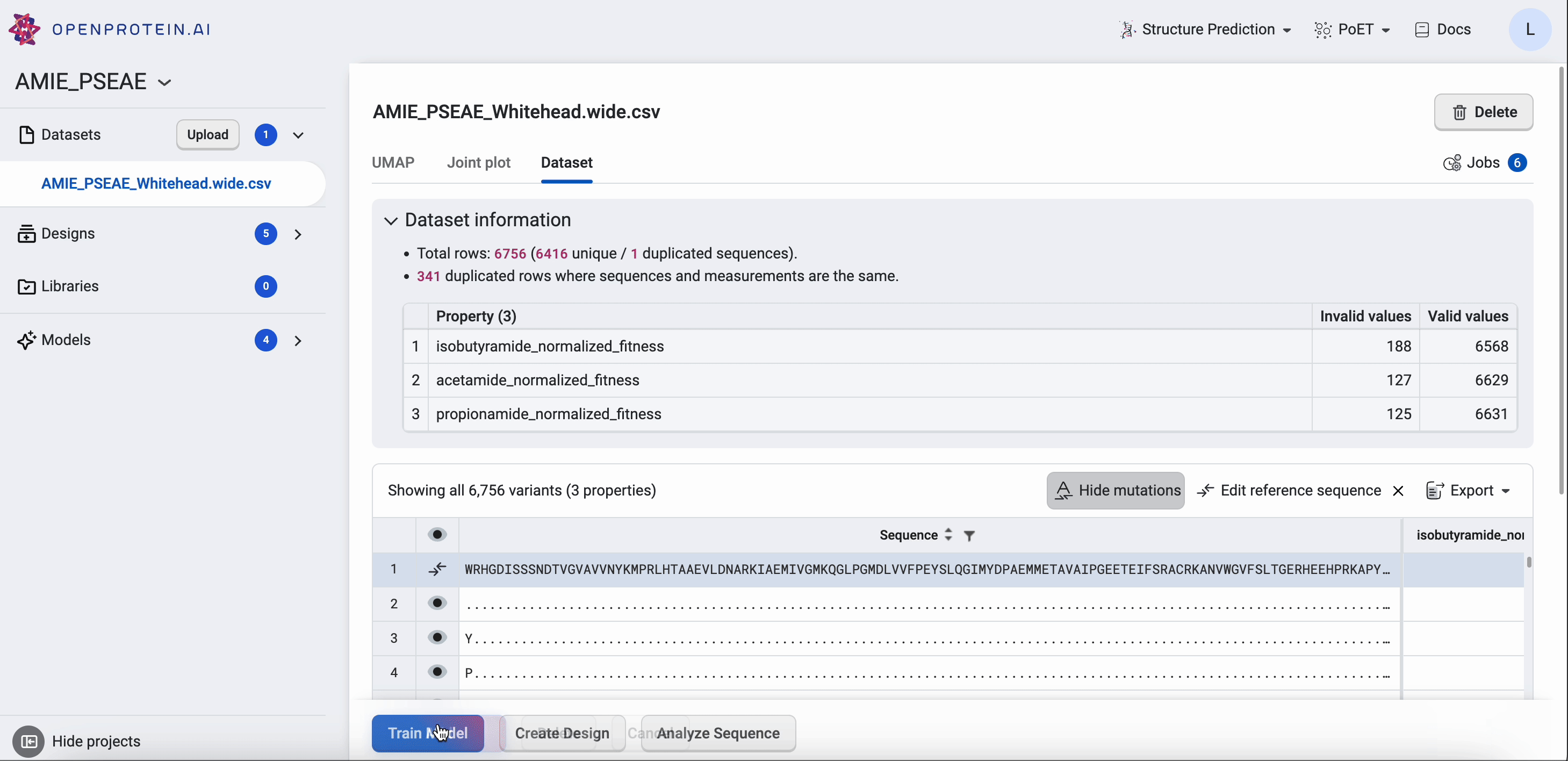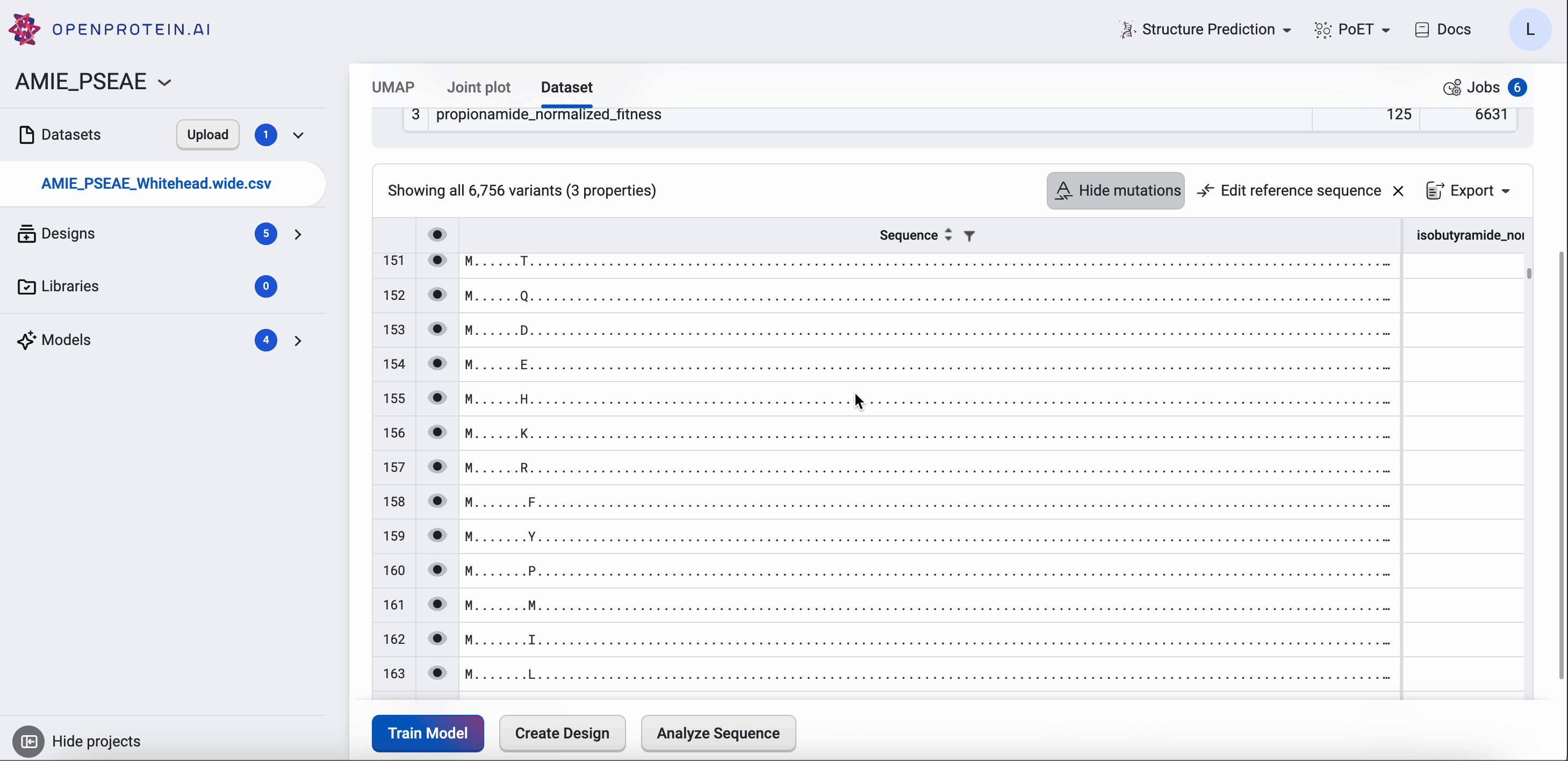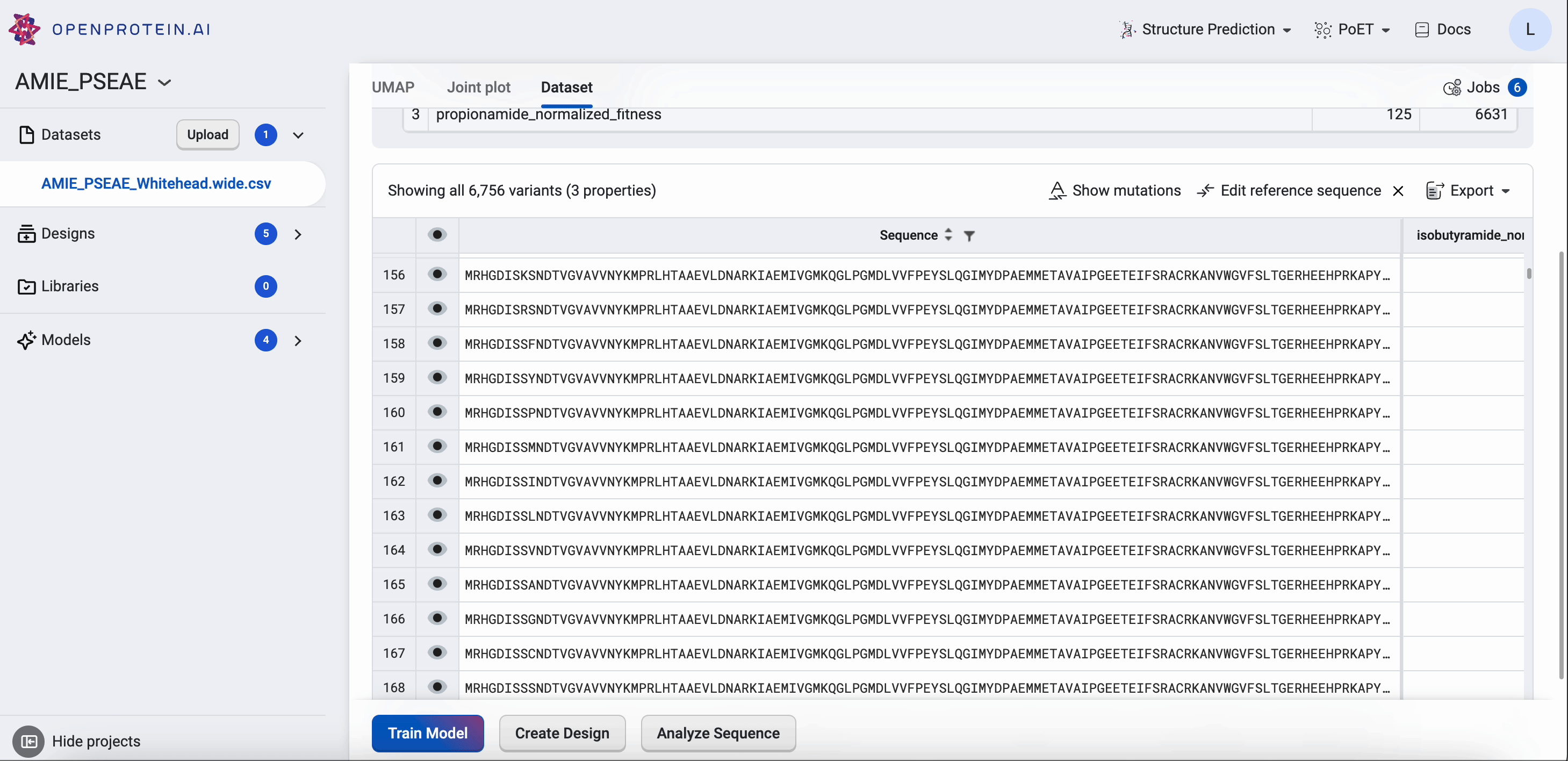
Your Dataset page shows the properties of your dataset, along with a table that you can filter and sort. If you want to compare your generated results against a benchmark, select Add a reference sequence , enter a parent sequence or sequence of interest, and select Add. You can update or delete reference sequences by selecting Edit reference sequence, choosing your desired action, then selecting Update. Using a reference sequence lets you view mutation sites to better understand specific substitutions present in your sequence libraries and designs.
Find more information about OpenProtein.AI’s visualization options in Visualizing your data.