Screening insertion locations with PoET#
In this tutorial, we’ll use PoET to evaluate the acceptibility of insertion locations for the nuclear localization signal PAAKRVKLD from c-Myc into a target protein.
[1]:
%matplotlib inline
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import json
[2]:
import openprotein
import openprotein.fasta as fasta
Connect to the OpenProtein.AI web server#
[3]:
with open('secrets.config', 'r') as f:
config = json.load(f)
session = openprotein.connect(config['username'], config['password'])
Define the peptide to insert and the protein to analyze#
We’ll examine insertion of the nuclear localization signal peptide from c-Myc.
[4]:
nls = b'PAAKRVKLD'
Examine insertion sites in aliphatic amidase#
[5]:
name = 'AMIE_PSEAE'
wt = b'MRHGDISSSNDTVGVAVVNYKMPRLHTAAEVLDNARKIAEMIVGMKQGLPGMDLVVFPEYSLQGIMYDPAEMMETAVAIPGEETEIFSRACRKANVWGVFSLTGERHEEHPRKAPYNTLVLIDNNGEIVQKYRKIIPWCPIEGWYPGGQTYVSEGPKGMKISLIICDDGNYPEIWRDCAMKGAELIVRCQGYMYPAKDQQVMMAKAMAWANNCYVAVANAAGFDGVYSYFGHSAIIGFDGRTLGECGEEEMGIQYAQLSLSQIRDARANDQSQNHLFKILHRGYSGLQASGDGDRGLAECPFEFYRTWVTDAEKARENVERLTRSTTGVAQCPVGRLPYEGLEKEA'
print('>' + name)
for i in range(0, len(wt), 80):
print(wt[i:i+80].decode())
>AMIE_PSEAE
MRHGDISSSNDTVGVAVVNYKMPRLHTAAEVLDNARKIAEMIVGMKQGLPGMDLVVFPEYSLQGIMYDPAEMMETAVAIP
GEETEIFSRACRKANVWGVFSLTGERHEEHPRKAPYNTLVLIDNNGEIVQKYRKIIPWCPIEGWYPGGQTYVSEGPKGMK
ISLIICDDGNYPEIWRDCAMKGAELIVRCQGYMYPAKDQQVMMAKAMAWANNCYVAVANAAGFDGVYSYFGHSAIIGFDG
RTLGECGEEEMGIQYAQLSLSQIRDARANDQSQNHLFKILHRGYSGLQASGDGDRGLAECPFEFYRTWVTDAEKARENVE
RLTRSTTGVAQCPVGRLPYEGLEKEA
Create the MSA and define an ensemble prompt#
To run PoET, we use the OpenProtein API to construct an MSA from homologues of the wildtype sequence using homology search and then define an ensemble prompt containing 10 prompts sampled with default parameters. Query sequences will then be scored conditioned on the fitness landscape represented by the sequences in the prompt.
OpenProtein uses an asynchronous API, where potentially long running functions return a job ID that can be used to query for completed results. The wait_until_done function can be used to poll for completion.
[6]:
# search for homologs to automatically create an MSA for the seed sequence
msa = session.align.create_msa(wt)
print(msa)
msa.wait_until_done(verbose=True)
# create the prompt, set the seed for reproducibility
prompt = msa.sample_prompt(num_ensemble_prompts=10, random_seed=1)
print(prompt)
prompt.wait_until_done(verbose=True)
job_id='73b0bdc0-b428-4f3a-a7d0-a1991a81b09a' job_type=<JobType.align_align: '/align/align'> status=<JobStatus.SUCCESS: 'SUCCESS'> created_date=datetime.datetime(2024, 10, 15, 16, 32, 37, 421115) start_date=None end_date=datetime.datetime(2024, 10, 15, 16, 32, 37, 421638) prerequisite_job_id=None progress_message=None progress_counter=None sequence_length=None
Waiting: 100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [00:00<00:00, 394.20it/s, status=SUCCESS]
job_id='463a1991-84c4-40b2-b373-6160f2695ce7' job_type=<JobType.align_prompt: '/align/prompt'> status=<JobStatus.PENDING: 'PENDING'> created_date=datetime.datetime(2024, 10, 15, 16, 32, 39, 291568) start_date=None end_date=None prerequisite_job_id=None progress_message=None progress_counter=None sequence_length=None
Waiting: 100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [00:21<00:00, 4.69it/s, status=SUCCESS]
[6]:
True
Enumerate and score the insertions#
[7]:
poet = session.embedding.get_model("poet")
[8]:
queries = [wt]
for i in range(len(wt)):
q = wt[:i] + nls + wt[i+1:]
queries.append(q)
len(queries)
[8]:
347
[9]:
future_scores = poet.score(prompt=prompt, sequences=queries)
print(future_scores.job)
future_scores.wait_until_done(verbose=True)
num_records=3470 job_id='88c791aa-8e0a-4c5d-9e7d-ddd89b5ba831' job_type=<JobType.poet_score: '/poet/score'> status=<JobStatus.PENDING: 'PENDING'> created_date=datetime.datetime(2024, 10, 15, 16, 33, 12, 119564, tzinfo=TzInfo(UTC)) start_date=None end_date=None prerequisite_job_id=None progress_message=None progress_counter=0 sequence_length=None
Waiting: 100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [00:42<00:00, 2.35it/s, status=SUCCESS]
[9]:
True
[10]:
results = future_scores.get()
scores_ensemble = np.stack([r[2] for r in results], axis=0)
scores_ensemble.shape
[10]:
(347, 10)
[11]:
baseline = scores_ensemble[0]
scores = scores_ensemble[1:].mean(axis=-1) - baseline.mean()
_, ax = plt.subplots(figsize=(20, 4))
ax.scatter(np.arange(1, len(wt)+1), scores)
ax.set_xlim(0, len(wt)+2)
plt.xlabel('Position')
plt.ylabel('PoET Likelihood Score')
plt.title('Insertion Site Tolerance Scan')
[11]:
Text(0.5, 1.0, 'Insertion Site Tolerance Scan')
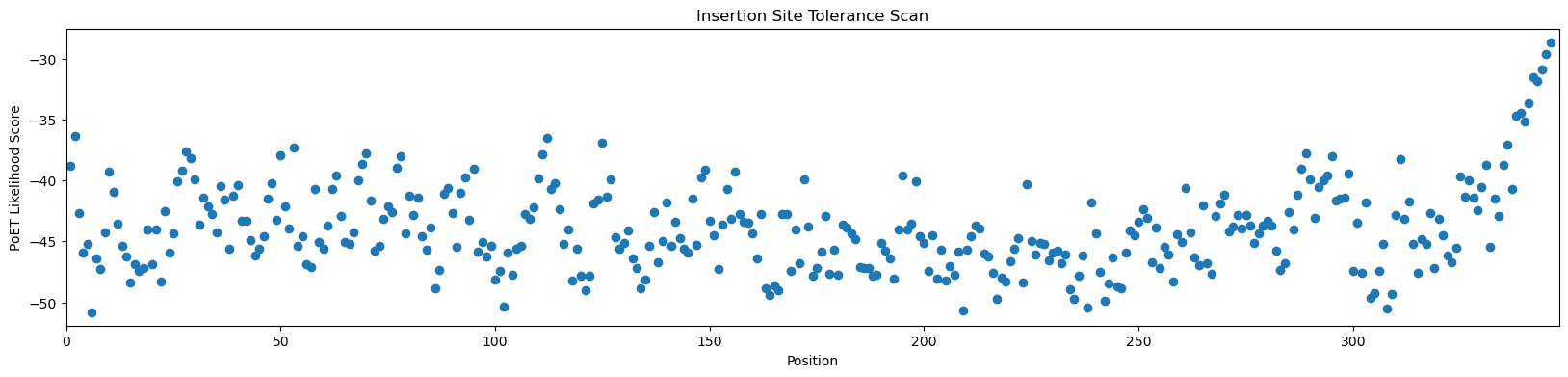
[12]:
# sort out the most favorable insertion sites
order = np.argsort(-scores)
print('Score ', 'Position', sep='\t')
for i in order[:10]:
print(f'{scores[i]:8.5f}', i+1, sep='\t')
Score Position
-29.84536 346
-31.18728 345
-31.35672 342
-32.17427 344
-32.20207 343
-32.69761 341
-33.74642 339
-34.27572 340
-34.92476 338
-36.01019 2
Sensibly, PoET predicts that the C-terminus is likely to be the most tolerated site to insert the NLS peptide. The N-terminus may also be tolerated.
Examine insertion sites in Cas9#
[13]:
from io import BytesIO
fasta_string = b'''>sp|J7RUA5|CAS9_STAAU CRISPR-associated endonuclease Cas9 OS=Staphylococcus aureus OX=1280 GN=cas9 PE=1 SV=1
MKRNYILGLDIGITSVGYGIIDYETRDVIDAGVRLFKEANVENNEGRRSKRGARRLKRRR
RHRIQRVKKLLFDYNLLTDHSELSGINPYEARVKGLSQKLSEEEFSAALLHLAKRRGVHN
VNEVEEDTGNELSTKEQISRNSKALEEKYVAELQLERLKKDGEVRGSINRFKTSDYVKEA
KQLLKVQKAYHQLDQSFIDTYIDLLETRRTYYEGPGEGSPFGWKDIKEWYEMLMGHCTYF
PEELRSVKYAYNADLYNALNDLNNLVITRDENEKLEYYEKFQIIENVFKQKKKPTLKQIA
KEILVNEEDIKGYRVTSTGKPEFTNLKVYHDIKDITARKEIIENAELLDQIAKILTIYQS
SEDIQEELTNLNSELTQEEIEQISNLKGYTGTHNLSLKAINLILDELWHTNDNQIAIFNR
LKLVPKKVDLSQQKEIPTTLVDDFILSPVVKRSFIQSIKVINAIIKKYGLPNDIIIELAR
EKNSKDAQKMINEMQKRNRQTNERIEEIIRTTGKENAKYLIEKIKLHDMQEGKCLYSLEA
IPLEDLLNNPFNYEVDHIIPRSVSFDNSFNNKVLVKQEENSKKGNRTPFQYLSSSDSKIS
YETFKKHILNLAKGKGRISKTKKEYLLEERDINRFSVQKDFINRNLVDTRYATRGLMNLL
RSYFRVNNLDVKVKSINGGFTSFLRRKWKFKKERNKGYKHHAEDALIIANADFIFKEWKK
LDKAKKVMENQMFEEKQAESMPEIETEQEYKEIFITPHQIKHIKDFKDYKYSHRVDKKPN
RELINDTLYSTRKDDKGNTLIVNNLNGLYDKDNDKLKKLINKSPEKLLMYHHDPQTYQKL
KLIMEQYGDEKNPLYKYYEETGNYLTKYSKKDNGPVIKKIKYYGNKLNAHLDITDDYPNS
RNKVVKLSLKPYRFDVYLDNGVYKFVTVKNLDVIKKENYYEVNSKCYEEAKKLKKISNQA
EFIASFYNNDLIKINGELYRVIGVNNDLLNRIEVNMIDITYREYLENMNDKRPPRIIKTI
ASKTQSIKKYSTDILGNLYEVKSKKHPQIIKKG'''
with BytesIO(fasta_string) as f:
names, sequences = fasta.parse(f)
name = names[0].decode()
wt = sequences[0]
print('>' + name)
for i in range(0, len(wt), 80):
print(wt[i:i+80].decode())
>sp|J7RUA5|CAS9_STAAU CRISPR-associated endonuclease Cas9 OS=Staphylococcus aureus OX=1280 GN=cas9 PE=1 SV=1
MKRNYILGLDIGITSVGYGIIDYETRDVIDAGVRLFKEANVENNEGRRSKRGARRLKRRRRHRIQRVKKLLFDYNLLTDH
SELSGINPYEARVKGLSQKLSEEEFSAALLHLAKRRGVHNVNEVEEDTGNELSTKEQISRNSKALEEKYVAELQLERLKK
DGEVRGSINRFKTSDYVKEAKQLLKVQKAYHQLDQSFIDTYIDLLETRRTYYEGPGEGSPFGWKDIKEWYEMLMGHCTYF
PEELRSVKYAYNADLYNALNDLNNLVITRDENEKLEYYEKFQIIENVFKQKKKPTLKQIAKEILVNEEDIKGYRVTSTGK
PEFTNLKVYHDIKDITARKEIIENAELLDQIAKILTIYQSSEDIQEELTNLNSELTQEEIEQISNLKGYTGTHNLSLKAI
NLILDELWHTNDNQIAIFNRLKLVPKKVDLSQQKEIPTTLVDDFILSPVVKRSFIQSIKVINAIIKKYGLPNDIIIELAR
EKNSKDAQKMINEMQKRNRQTNERIEEIIRTTGKENAKYLIEKIKLHDMQEGKCLYSLEAIPLEDLLNNPFNYEVDHIIP
RSVSFDNSFNNKVLVKQEENSKKGNRTPFQYLSSSDSKISYETFKKHILNLAKGKGRISKTKKEYLLEERDINRFSVQKD
FINRNLVDTRYATRGLMNLLRSYFRVNNLDVKVKSINGGFTSFLRRKWKFKKERNKGYKHHAEDALIIANADFIFKEWKK
LDKAKKVMENQMFEEKQAESMPEIETEQEYKEIFITPHQIKHIKDFKDYKYSHRVDKKPNRELINDTLYSTRKDDKGNTL
IVNNLNGLYDKDNDKLKKLINKSPEKLLMYHHDPQTYQKLKLIMEQYGDEKNPLYKYYEETGNYLTKYSKKDNGPVIKKI
KYYGNKLNAHLDITDDYPNSRNKVVKLSLKPYRFDVYLDNGVYKFVTVKNLDVIKKENYYEVNSKCYEEAKKLKKISNQA
EFIASFYNNDLIKINGELYRVIGVNNDLLNRIEVNMIDITYREYLENMNDKRPPRIIKTIASKTQSIKKYSTDILGNLYE
VKSKKHPQIIKKG
Create the MSA and define an ensemble prompt#
[14]:
# search for homologs to automatically create an MSA for the seed sequence
msa = session.align.create_msa(wt)
print(msa)
msa.wait_until_done(verbose=True)
# create the prompt, set the seed for reproducibility
prompt = msa.sample_prompt(num_ensemble_prompts=10, random_seed=1)
print(prompt)
prompt.wait_until_done(verbose=True)
job_id='40c78574-3dbe-4b80-9a42-6dae4c27ab16' job_type=<JobType.align_align: '/align/align'> status=<JobStatus.SUCCESS: 'SUCCESS'> created_date=datetime.datetime(2024, 10, 15, 16, 34, 6, 334299) start_date=None end_date=datetime.datetime(2024, 10, 15, 16, 34, 6, 334769) prerequisite_job_id=None progress_message=None progress_counter=None sequence_length=None
Waiting: 100%|██████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [00:00<00:00, 351.38it/s, status=SUCCESS]
job_id='22a07da8-0591-49ad-aad5-040284c03999' job_type=<JobType.align_prompt: '/align/prompt'> status=<JobStatus.PENDING: 'PENDING'> created_date=datetime.datetime(2024, 10, 15, 16, 34, 8, 187496) start_date=None end_date=None prerequisite_job_id=None progress_message=None progress_counter=None sequence_length=None
Waiting: 100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [01:57<00:00, 1.18s/it, status=SUCCESS]
[14]:
True
Enumerate and score the insertions#
[15]:
queries = [wt]
for i in range(len(wt)):
q = wt[:i] + nls + wt[i+1:]
queries.append(q)
len(queries)
[15]:
1054
[16]:
future_scores = poet.score(prompt=prompt, sequences=queries)
print(future_scores.job)
future_scores.wait_until_done(verbose=True)
num_records=10540 job_id='a65c4ebb-b0d5-4e8d-84cb-6026f680dd67' job_type=<JobType.poet_score: '/poet/score'> status=<JobStatus.PENDING: 'PENDING'> created_date=datetime.datetime(2024, 10, 15, 16, 36, 15, 501964, tzinfo=TzInfo(UTC)) start_date=None end_date=None prerequisite_job_id=None progress_message=None progress_counter=0 sequence_length=None
Waiting: 100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [01:56<00:00, 1.17s/it, status=SUCCESS]
[16]:
True
[17]:
results = future_scores.get()
scores_ensemble = np.stack([r[2] for r in results], axis=0)
scores_ensemble.shape
[17]:
(1054, 10)
[18]:
baseline = scores_ensemble[0]
scores = scores_ensemble[1:].mean(axis=-1) - baseline.mean()
_, ax = plt.subplots(figsize=(20, 4))
ax.scatter(np.arange(1, len(wt)+1), scores)
ax.set_xlim(0, len(wt)+2)
plt.xlabel('Position')
plt.ylabel('PoET Likelihood Score')
plt.title('Insertion Site Tolerance Scan')
[18]:
Text(0.5, 1.0, 'Insertion Site Tolerance Scan')
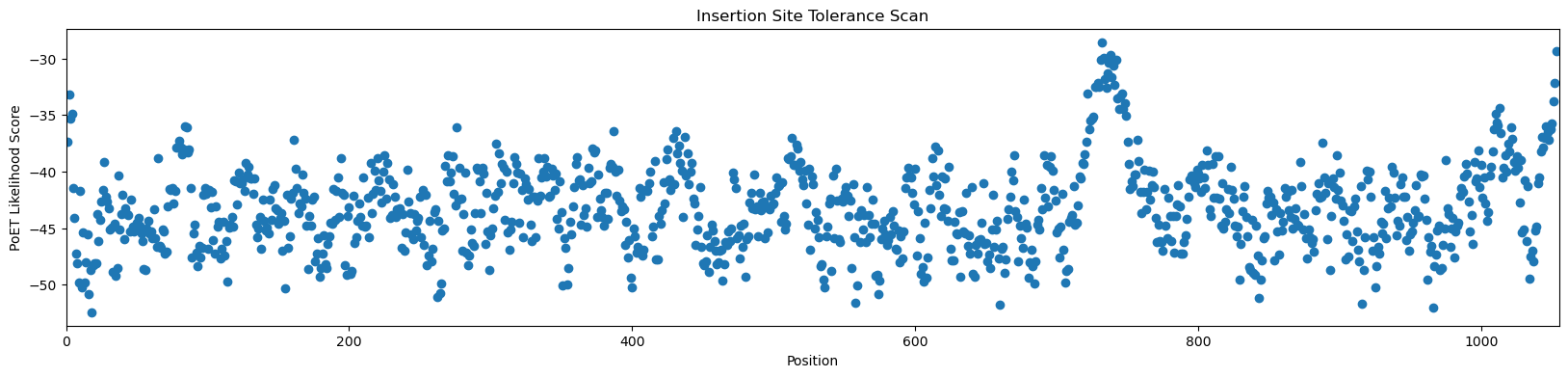
[19]:
# sort out the most favorable insertion sites
order = np.argsort(-scores)
print('Score ', 'Position', sep='\t')
for i in order[:10]:
print(f'{scores[i]:8.5f}', i+1, sep='\t')
Score Position
-28.55595 732
-29.33330 1053
-29.71844 738
-29.94032 733
-30.09066 742
-30.16471 731
-30.34108 737
-30.61783 740
-31.26478 736
-31.68063 739
For Cas9, PoET predicts that the best tolerated insertion site is at position 732 or position 738. The C-terminus is also predited to be amongst the best tolerated. Examining the plot also shows that the N-terminus should also be tolerated. Engineered Cas9 proteins have used both N- and C-terminal nuclear localization signals, but the internal 732/738 sites are novel predictions.
Examining the crystal structure of this Cas9, 5AXW, shows that the the 732-738 stretch is a surface exposed loop, a plausible insertion location.